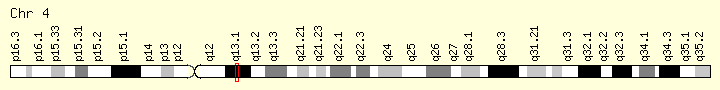ADGRL3 (adhesion G protein-coupled receptor L3)
- symbol:
- ADGRL3
- locus group:
- protein-coding gene
- location:
- 4q13.1
- gene_family:
- Adhesion G protein-coupled receptors, subfamily L
- alias symbol:
- KIAA0768|LEC3
- alias name:
- None
- entrez id:
- 23284
- ensembl gene id:
- ENSG00000150471
- ucsc gene id:
- None
- refseq accession:
- NM_001322246
- hgnc_id:
- HGNC:20974
- approved reserved:
- 2003-04-25
ADGRL3(也称为LPHN3)属于粘附G蛋白偶联受体(aGPCR)家族,是一种在神经系统中高度表达的跨膜蛋白受体。它的主要功能是通过与细胞外基质蛋白(如teneurins和latrophilins)相互作用参与突触形成和神经元通讯,对大脑发育、突触可塑性和神经递质释放调节起关键作用。ADGRL3在注意力缺陷多动障碍(ADHD)和物质成瘾等神经精神疾病中具有重要关联,全基因组关联研究(GWAS)发现其特定单核苷酸多态性(SNP)与ADHD风险显著相关。该基因突变可能导致受体构象变化或信号转导异常,进而影响突触功能并增加行为障碍易感性。ADGRL3过表达会增强神经元粘附和突触稳定性,但可能降低神经回路可塑性;而表达降低则导致突触形成缺陷、多巴胺信号失调,与冲动行为和注意力不集中相关。作为aGPCR家族成员,ADGRL3具有典型的结构特征:包含GPS(GPCR蛋白水解位点)结构域,可发生自剪切产生N端和C端片段,并通过Stachel序列激活下游G蛋白信号通路。该家族共性在于通过机械力或配体结合触发胞内信号,参与细胞粘附、迁移和组织发育等过程。ADGRL3还与FLRT3等蛋白形成跨突触复合物,其表达异常可能影响整个神经网络连接性,特别是在前额叶皮层和纹状体等与执行功能相关的脑区。
This gene encodes a member of the latrophilin subfamily of G-protein coupled receptors (GPCR). Latrophilins may function in both cell adhesion and signal transduction. In experiments with non-human species, endogenous proteolytic cleavage within a cysteine-rich GPS (G-protein-coupled-receptor proteolysis site) domain resulted in two subunits (a large extracellular N-terminal cell adhesion subunit and a subunit with substantial similarity to the secretin/calcitonin family of GPCRs) being non-covalently bound at the cell membrane. [provided by RefSeq, Jul 2008]
这个基因编码的G蛋白偶联受体(GPCR)的latrophilin亚科的成员。 Latrophilins可以在两个细胞粘附和信号转导的作用。在具有富含半胱氨酸的GPS内的非人类物种内源性蛋白水解切割(G蛋白偶联受体蛋白水解位点)的实验域导致两个亚基(一个大的细胞外N-末端细胞粘附亚基和与实质相似的子单元GPCR的胰液/降钙素家族)被在细胞膜非共价结合。 [由RefSeq的,2008年7月提供]
基因本体信息
ADGRL3基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
ADGRL3基因的本体(GO)信息:
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。