CDK7 (cyclin dependent kinase 7)
- symbol:
- CDK7
- locus group:
- protein-coding gene
- location:
- 5q13.2
- gene_family:
- Cyclin-dependent kinases
- alias symbol:
- CAK1|CDKN7|MO15|STK1|CAK
- alias name:
- None
- entrez id:
- 1022
- ensembl gene id:
- ENSG00000134058
- ucsc gene id:
- uc003jvs.5
- refseq accession:
- NM_001799
- hgnc_id:
- HGNC:1778
- approved reserved:
- 1994-12-16
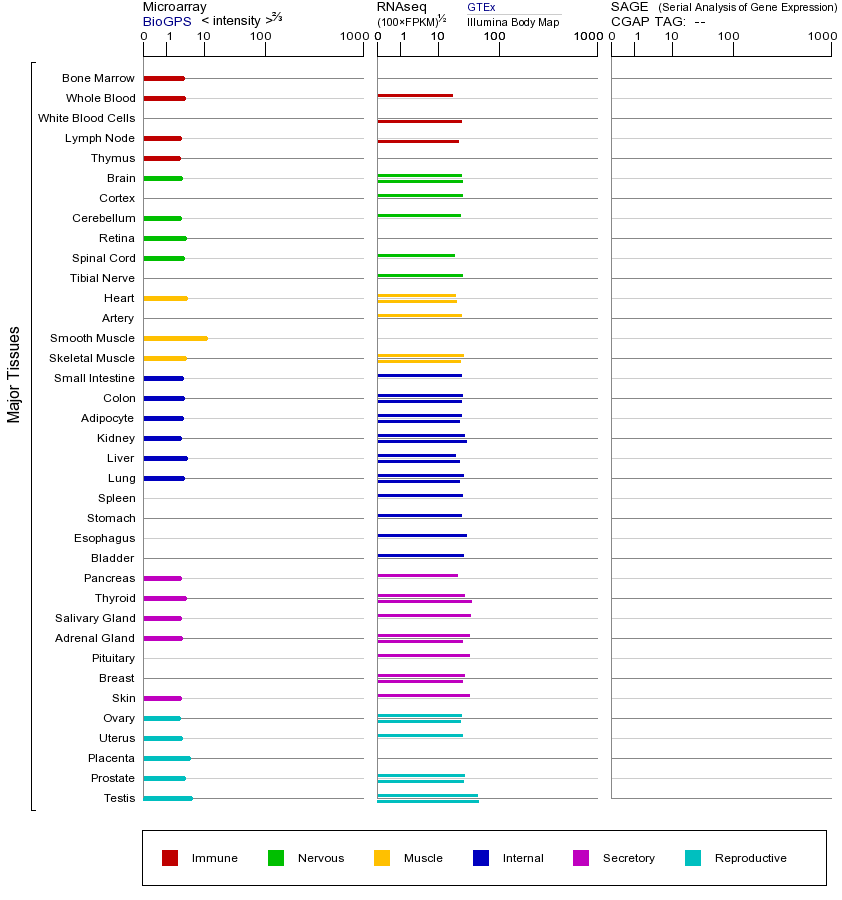
CDK7(细胞周期蛋白依赖性激酶7)是一个关键的蛋白激酶,属于CDK基因家族,该家族成员普遍参与细胞周期调控和转录调节。CDK7的主要功能是作为细胞周期调控的核心组分,同时也是转录因子TFIIH的组成部分,负责RNA聚合酶II的羧基末端结构域(CTD)磷酸化,从而促进转录起始。此外,CDK7还能激活其他CDKs(如CDK1、CDK2、CDK4和CDK6),通过磷酸化它们的T-loop区域来调控细胞周期进程。CDK7在多种组织中广泛表达,尤其在增殖活跃的细胞中表现显著。突变或异常表达可能严重影响细胞周期和转录,导致疾病发生。例如,CDK7功能丧失突变可能导致发育缺陷或细胞周期停滞,而过表达则与多种癌症(如乳腺癌、肺癌和卵巢癌)的发生密切相关,因其促进细胞异常增殖和存活。CDK7的抑制已被探索为癌症治疗策略,例如使用THZ1等特异性抑制剂靶向CDK7的ATP结合位点,可有效抑制肿瘤生长。此外,CDK7还参与DNA损伤修复,其功能异常可能导致基因组不稳定。在基因家族层面,CDKs的共同特点是依赖与细胞周期蛋白(cyclin)结合来发挥激酶活性,调控细胞周期过渡或转录过程。CDK7的过表达通常加速细胞周期进程并增强转录活性,可能引发癌变;而表达降低则可能导致细胞周期阻滞或转录缺陷,影响正常细胞功能。研究还发现CDK7与其他基因(如MYC和ERα)存在功能交互,进一步放大其在肿瘤发生中的作用。
The protein encoded by this gene is a member of the cyclin-dependent protein kinase (CDK) family. CDK family members are highly similar to the gene products of Saccharomyces cerevisiae cdc28, and Schizosaccharomyces pombe cdc2, and are known to be important regulators of cell cycle progression. This protein forms a trimeric complex with cyclin H and MAT1, which functions as a Cdk-activating kinase (CAK). It is an essential component of the transcription factor TFIIH, that is involved in transcription initiation and DNA repair. This protein is thought to serve as a direct link between the regulation of transcription and the cell cycle. [provided by RefSeq, Jul 2008]
由该基因编码的蛋白质是细胞周期蛋白依赖性蛋白激酶(CDK)家族的一个成员。 CDK家族成员是高度相似的酿酒酵母CDC28基因产物,和粟酒裂殖酵母CDC2,并已知是细胞周期进展的重要调节剂。此蛋白与细胞周期蛋白H和MAT1,其功能是作为一个CDK激活激酶(CAK)三聚体复合物。它是转录因子TFIIH,是参与转录起始和DNA修复的一个重要组成部分。这种蛋白质被认为是作为转录调控和细胞周期之间的直接联系。 [由RefSeq的,2008年7月提供]
基因本体信息
CDK7基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
CDK7基因的本体(GO)信息:
| 名称 |
|---|
| 3022 Basal transcription factors [PATH:hsa03022] |
| 3420 Nucleotide excision repair [PATH:hsa03420] |
| 4110 Cell cycle [PATH:hsa04110] |
| 名称 |
|---|
| Cell Cycle |
| Cell Cycle, Mitotic |
| Cyclin A:Cdk2-associated events at S phase entry |
| Cyclin A/B1 associated events during G2/M transition |
| Cyclin D associated events in G1 |
| Cyclin E associated events during G1/S transition |
| Disease |
| DNA Repair |
| Dual incision reaction in GG-NER |
| Dual incision reaction in TC-NER |
| Epigenetic regulation of gene expression |
| Formation of HIV elongation complex in the absence of HIV Tat |
| Formation of HIV-1 elongation complex containing HIV-1 Tat |
| Formation of incision complex in GG-NER |
| Formation of RNA Pol II elongation complex |
| Formation of the Early Elongation Complex |
| Formation of the HIV-1 Early Elongation Complex |
| Formation of transcription-coupled NER (TC-NER) repair complex |
| G1 Phase |
| G1/S Transition |
| G2/M Transition |
| Gene Expression |
| Global Genomic NER (GG-NER) |
| HIV Infection |
| HIV Life Cycle |
| HIV Transcription Elongation |
| HIV Transcription Initiation |
| Infectious disease |
| Late Phase of HIV Life Cycle |
| Mitotic G1-G1/S phases |
| Mitotic G2-G2/M phases |
| mRNA Capping |
| Negative epigenetic regulation of rRNA expression |
| NoRC negatively regulates rRNA expression |
| Nucleotide Excision Repair |
| RNA Pol II CTD phosphorylation and interaction with CE |
| RNA Polymerase I Chain Elongation |
| RNA Polymerase I Promoter Clearance |
| RNA Polymerase I Promoter Escape |
| RNA Polymerase I Transcription |
| RNA Polymerase I Transcription Initiation |
| RNA Polymerase I Transcription Termination |
| RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| RNA Polymerase II HIV Promoter Escape |
| RNA Polymerase II Pre-transcription Events |
| RNA Polymerase II Promoter Escape |
| RNA Polymerase II Transcription |
| RNA Polymerase II Transcription Elongation |
| RNA Polymerase II Transcription Initiation |
| RNA Polymerase II Transcription Initiation And Promoter Clearance |
| RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| S Phase |
| Tat-mediated elongation of the HIV-1 transcript |
| Transcription |
| Transcription of the HIV genome |
| Transcription-coupled NER (TC-NER) |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Peripheral Neuropathy | 0.12 | 1 | 0 | CTD_human |
| Malignant neoplasm of breast | 0.005005506 | 2 | 0 | BeFree_GAD |
| Lung Neoplasms | 0.00272435 | 1 | 0 | LHGDN |
| Glioma | 0.00272435 | 1 | 0 | LHGDN |
| leukemia | 0.002638474 | 2 | 0 | BeFree_GAD |
| Malignant neoplasm of lung | 0.002638474 | 2 | 0 | BeFree_GAD |
| Malignant neoplasm of ovary | 0.002638474 | 1 | 0 | BeFree_GAD |
| Rectal Neoplasms | 0.002367032 | 1 | 0 | GAD |
| Diabetes Mellitus, Non-Insulin-Dependent | 0.002367032 | 1 | 0 | GAD |
| body mass | 0.002367032 | 1 | 0 | GAD |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。