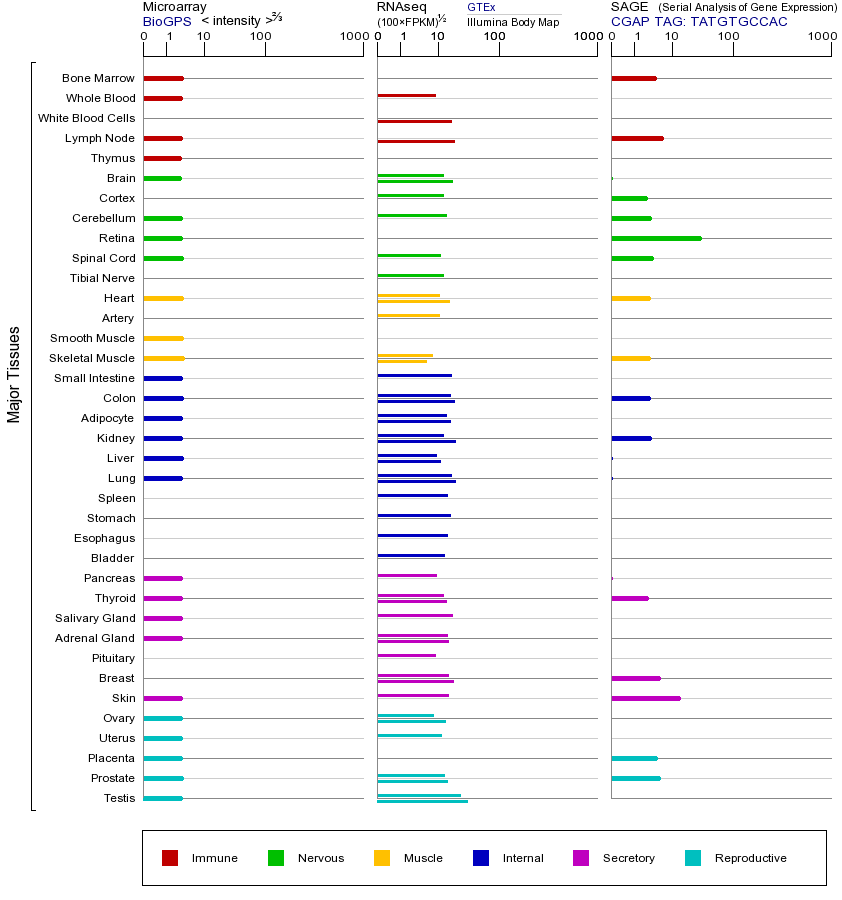
ECT2 (epithelial cell transforming 2)
- symbol:
- ECT2
- locus group:
- protein-coding gene
- location:
- 3q26.31
- gene_family:
- Rho guanine nucleotide exchange factors
- alias symbol:
- ARHGEF31
- alias name:
- None
- entrez id:
- 1894
- ensembl gene id:
- ENSG00000114346
- ucsc gene id:
- uc003fii.4
- refseq accession:
- NM_018098
- hgnc_id:
- HGNC:3155
- approved reserved:
- 1993-10-15
ECT2(上皮细胞转化序列2)是一种属于Rho鸟苷酸交换因子(RhoGEF)家族的基因,该家族成员通过激活Rho GTPases(如RhoA、Rac1和Cdc42)调控细胞骨架重组、细胞迁移和分裂等关键生物学过程。ECT2主要在细胞质中发挥作用,尤其在细胞有丝分裂期间,它通过激活RhoA信号通路促进胞质分裂环的形成,确保细胞正常分裂。ECT2的突变或异常表达会破坏这一过程,导致染色体不稳定或多核细胞形成,进而可能促进肿瘤发生。研究表明,ECT2在多种癌症(如肺癌、乳腺癌和胶质瘤)中过表达,其高表达水平与肿瘤侵袭性增强、预后不良相关,这可能是因为它过度激活Rho信号通路,促进癌细胞迁移和转移。相反,降低ECT2表达会抑制肿瘤细胞增殖和转移能力,但可能干扰正常细胞的胞质分裂。ECT2还参与组织修复和胚胎发育等生理过程。该基因家族(RhoGEF)的共性是通过催化Rho GTPases的GDP/GTP交换,调控下游信号通路,影响细胞形态、运动和增殖。ECT2的致癌特性使其成为潜在的治疗靶点,针对其过度活化的抑制剂可能具有抗肿瘤效果。
The protein encoded by this gene is a guanine nucleotide exchange factor and transforming protein that is related to Rho-specific exchange factors and yeast cell cycle regulators. The expression of this gene is elevated with the onset of DNA synthesis and remains elevated during G2 and M phases. In situ hybridization analysis showed that expression is at a high level in cells undergoing mitosis in regenerating liver. Thus, this protein is expressed in a cell cycle-dependent manner during liver regeneration, and is thought to have an important role in the regulation of cytokinesis. Several transcript variants encoding two different isoforms have been found for this gene. [provided by RefSeq, Apr 2012]
由该基因编码的蛋白质是一个鸟嘌呤核苷酸交换因子和转化蛋白质是相关的Rho-特定交换因子和酵母细胞周期调节。该基因的表达升高与DNA合成的起始和G2和M期期间保持升高。原位杂交分析表明,表达是在在再生肝经历有丝分裂的细胞的高的水平。因此,该蛋白在肝再生过程中的细胞周期依赖性的方式表达,并且被认为具有在胞质分裂的调节中起重要作用。一些转录变异体的编码两个不同的亚型已经发现了这种基因。 [由RefSeq的,2012年4月提供]
基因本体信息
ECT2基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
ECT2基因的本体(GO)信息:
| 名称 |
|---|
| Cell death signalling via NRAGE, NRIF and NADE |
| G alpha (12/13) signalling events |
| GPCR downstream signaling |
| NRAGE signals death through JNK |
| p75 NTR receptor-mediated signalling |
| Rho GTPase cycle |
| Signal Transduction |
| Signaling by GPCR |
| Signaling by Rho GTPases |
| Signalling by NGF |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| IGA Glomerulonephritis | 0.12 | 1 | 0 | CTD_human |
| Glioblastoma | 0.003267234 | 2 | 0 | BeFree_LHGDN |
| Pancreatic Neoplasm | 0.002995792 | 1 | 0 | BeFree_LHGDN |
| Pancreatic carcinoma | 0.000542884 | 2 | 0 | BeFree |
| Adenocarcinoma | 0.000542884 | 2 | 0 | BeFree |
| Malignant neoplasm of pancreas | 0.000542884 | 2 | 0 | BeFree |
| Glioma | 0.000542884 | 2 | 0 | BeFree |
| Osteosarcoma of bone | 0.000542884 | 2 | 0 | BeFree |
| Non-Small Cell Lung Carcinoma | 0.000542884 | 2 | 0 | BeFree |
| Osteosarcoma | 0.000542884 | 2 | 0 | BeFree |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。