FAS (Fas cell surface death receptor)
- symbol:
- FAS
- locus group:
- protein-coding gene
- location:
- 10q23.31
- gene_family:
- CD molecules|Tumor necrosis factor receptor superfamily
- alias symbol:
- CD95|APO-1
- alias name:
- TNF receptor superfamily member 6
- entrez id:
- 355
- ensembl gene id:
- ENSG00000026103
- ucsc gene id:
- uc001kfr.4
- refseq accession:
- NM_000043
- hgnc_id:
- HGNC:11920
- approved reserved:
- 1992-06-25
FAS基因(又称FAS细胞表面死亡受体或TNFRSF6)属于肿瘤坏死因子受体超家族(TNF receptor superfamily),主要编码一种称为Fas受体(FasR)的跨膜蛋白。Fas受体与其配体FasL(Fas ligand)结合后,可激活细胞凋亡(程序性细胞死亡)信号通路,在免疫调节、清除异常细胞及维持组织稳态中起关键作用。该基因的主要作用位点为细胞膜,其激活后通过胞内死亡结构域(death domain)招募FADD(Fas-associated protein with death domain),进而启动caspase级联反应,最终导致细胞凋亡。FAS基因突变可能导致功能丧失,引发自身免疫性淋巴增生综合征(ALPS),表现为淋巴细胞异常积累、脾肿大及自身免疫疾病;而过度激活可能与肝细胞凋亡相关的肝炎或组织损伤有关。FAS与多种疾病相关,例如癌症中其表达下调可能导致肿瘤细胞逃避免疫清除,而在神经退行性疾病中异常激活可能加剧神经元凋亡。若FAS过表达,可能引起过度细胞凋亡,导致组织萎缩或器官功能障碍;若表达降低,则可能抑制正常凋亡过程,促进自身免疫或肿瘤发展。FAS基因家族(TNFRSF)的共性包括:均含有富含半胱氨酸的胞外结构域(cysteine-rich domain)和胞内死亡结构域,多数成员通过调控细胞存活或凋亡参与免疫应答。专业术语解释:细胞凋亡(apoptosis)是受控的细胞自我清除过程;死亡结构域(death domain)是介导凋亡信号传递的蛋白相互作用模块;caspase是一类促进凋亡的蛋白酶。若译为“Fas配体”可能更准确(原英文:Fas ligand)。
The protein encoded by this gene is a member of the TNF-receptor superfamily. This receptor contains a death domain. It has been shown to play a central role in the physiological regulation of programmed cell death, and has been implicated in the pathogenesis of various malignancies and diseases of the immune system. The interaction of this receptor with its ligand allows the formation of a death-inducing signaling complex that includes Fas-associated death domain protein (FADD), caspase 8, and caspase 10. The autoproteolytic processing of the caspases in the complex triggers a downstream caspase cascade, and leads to apoptosis. This receptor has been also shown to activate NF-kappaB, MAPK3/ERK1, and MAPK8/JNK, and is found to be involved in transducing the proliferating signals in normal diploid fibroblast and T cells. Several alternatively spliced transcript variants have been described, some of which are candidates for nonsense-mediated mRNA decay (NMD). The isoforms lacking the transmembrane domain may negatively regulate the apoptosis mediated by the full length isoform. [provided by RefSeq, Mar 2011]
由该基因编码的蛋白质是TNF受体超家族的成员。这种受体含有死亡域。它已被显示出在程序性细胞死亡的生理调节中心作用,并在各种恶性肿瘤和免疫系统疾病的发病机理有牵连。该受体与其配体的相互作用可以包括Fas相关死亡域蛋白(FADD),胱天蛋白酶8,和caspase 10.死亡诱导信号传导复合物的形成的半胱氨酸蛋白酶,在复杂的自我蛋白酶处理触发下游胱天蛋白酶级联,并导致细胞凋亡。该受体也已显示活化的NF-κB的,MAPK3 / ERK1和MAPK8 / JNK和被发现涉及在正常二倍体成纤维细胞和T细胞转导的增殖信号。几个可变剪接转录物变体已有描述,其中一些是对无义介导的mRNA降解(NMD)的候选者。缺少跨膜结构域的同种型可通过全长同种型介导的细胞凋亡负调控。 [由RefSeq的,2011年3月提供]
基因本体信息
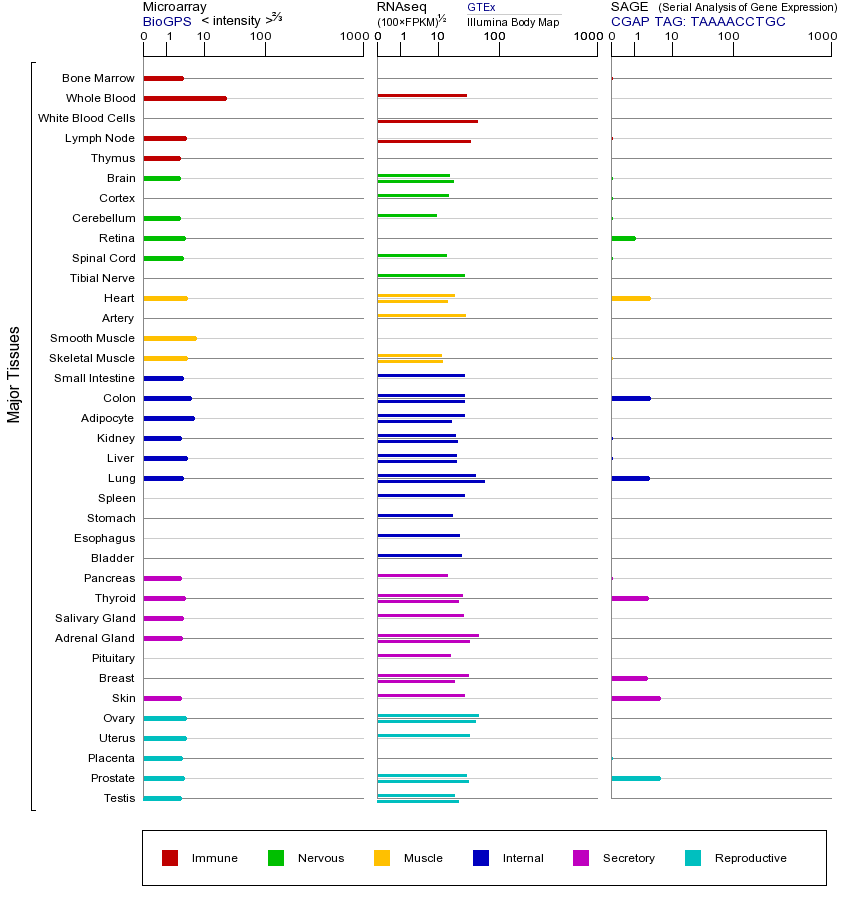
FAS基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
FAS基因的本体(GO)信息:
| 名称 |
|---|
| 4010 MAPK signaling pathway [PATH:hsa04010] |
| 4668 TNF signaling pathway [PATH:hsa04668] |
| 4060 Cytokine-cytokine receptor interaction [PATH:hsa04060] |
| 4210 Apoptosis [PATH:hsa04210] |
| 4115 p53 signaling pathway [PATH:hsa04115] |
| 4650 Natural killer cell mediated cytotoxicity [PATH:hsa04650] |
| 5200 Pathways in cancer [PATH:hsa05200] |
| 5205 Proteoglycans in cancer [PATH:hsa05205] |
| 5320 Autoimmune thyroid disease [PATH:hsa05320] |
| 5330 Allograft rejection [PATH:hsa05330] |
| 5332 Graft-versus-host disease [PATH:hsa05332] |
| 5010 Alzheimer's disease [PATH:hsa05010] |
| 4940 Type I diabetes mellitus [PATH:hsa04940] |
| 4932 Non-alcoholic fatty liver disease (NAFLD) [PATH:hsa04932] |
| 5162 Measles [PATH:hsa05162] |
| 5164 Influenza A [PATH:hsa05164] |
| 5161 Hepatitis B [PATH:hsa05161] |
| 5168 Herpes simplex infection [PATH:hsa05168] |
| 5142 Chagas disease (American trypanosomiasis) [PATH:hsa05142] |
| 5143 African trypanosomiasis [PATH:hsa05143] |
| 名称 |
|---|
| Apoptosis |
| CASP8 activity is inhibited |
| Caspase activation via extrinsic apoptotic signalig pathway |
| Death Receptor Signalling |
| Dimerization of procaspase-8 |
| FasL/ CD95L signaling |
| Ligand-dependent caspase activation |
| Programmed Cell Death |
| Regulated Necrosis |
| Regulation by c-FLIP |
| Regulation of necroptotic cell death |
| RIPK1-mediated regulated necrosis |
| Signal Transduction |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Autoimmune Lymphoproliferative Syndrome | 0.462725172 | 81 | 1 | BeFree_CTD_human_GAD_MGD_ORPHANET_UNIPROT |
| Cardiomyopathy, Dilated | 0.200271442 | 3 | 0 | BeFree_CTD_human_RGD |
| Diabetes Mellitus, Experimental | 0.2 | 2 | 0 | CTD_human_RGD |
| Lung Neoplasms | 0.133893193 | 7 | 0 | BeFree_CTD_human_LHGDN |
| Autoimmune Diseases | 0.130801593 | 16 | 0 | BeFree_CTD_human_GAD_LHGDN |
| Chronic Lymphocytic Leukemia | 0.127891677 | 12 | 2 | BeFree_GWASCAT_LHGDN |
| Leukemia, Myelocytic, Acute | 0.127177041 | 10 | 0 | BeFree_CTD_human_GAD |
| Liver Failure, Acute | 0.123267234 | 4 | 0 | BeFree_CTD_human_LHGDN |
| Diabetes Mellitus, Non-Insulin-Dependent | 0.122638474 | 3 | 0 | BeFree_CTD_human_GAD |
| Lymphoma, T-Cell, Cutaneous | 0.121628651 | 7 | 0 | BeFree_CTD_human |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。