LHCGR (luteinizing hormone/choriogonadotropin receptor)
- symbol:
- LHCGR
- locus group:
- protein-coding gene
- location:
- 2p16.3
- gene_family:
- Glycoprotein hormone receptors
- alias symbol:
- LHR|LCGR|LGR2|ULG5
- alias name:
- luteinizing hormone receptor
- entrez id:
- 3973
- ensembl gene id:
- ENSG00000138039
- ucsc gene id:
- uc002rwu.5
- refseq accession:
- NM_000233.3
- hgnc_id:
- HGNC:6585
- approved reserved:
- 1990-03-05
LHCGR(黄体生成素/绒毛膜促性腺激素受体)是一种G蛋白偶联受体,属于糖蛋白激素受体家族,主要表达于卵巢和睾丸中。该基因编码的蛋白质能够结合黄体生成素(LH)和人绒毛膜促性腺激素(hCG),激活下游信号通路(如cAMP/PKA),调控性腺功能,包括促进睾酮合成、卵泡发育、排卵和黄体形成。LHCGR的突变可能导致多种疾病,如男性假两性畸形(Leydig细胞发育不全)、女性原发性闭经或卵巢早衰。功能丧失性突变会破坏激素信号传导,导致性腺功能减退;而功能获得性突变可能引发家族性男性性早熟。LHCGR过表达可能与多囊卵巢综合征(PCOS)和某些卵巢肿瘤相关,因其过度激活会增强性激素生成;表达降低则可能导致不育或性腺功能低下。该基因属于糖蛋白激素受体家族,成员还包括FSHR(卵泡刺激素受体)和TSHR(促甲状腺激素受体),它们均具有相似的七次跨膜结构,通过G蛋白介导信号转导,参与调控生殖和内分泌功能。
This gene encodes the receptor for both luteinizing hormone and choriogonadotropin. This receptor belongs to the G-protein coupled receptor 1 family, and its activity is mediated by G proteins which activate adenylate cyclase. Mutations in this gene result in disorders of male secondary sexual character development, including familial male precocious puberty, also known as testotoxicosis, hypogonadotropic hypogonadism, Leydig cell adenoma with precocious puberty, and male pseudohermaphtoditism with Leydig cell hypoplasia. [provided by RefSeq, Jul 2008]
该基因编码的促黄体激素和绒毛膜促性腺激素两个受体。这种受体属于G-蛋白偶联受体1家族,其活性是由激活腺苷酸环化酶的G蛋白介导的。突变这个基因导致男性第二性征发育,包括家族男性性早熟,也被称为testotoxicosis,性腺机能减退,睾丸间质细胞腺瘤性早熟,和男性pseudohermaphtoditism与间质细胞发育不全的疾病。 [由RefSeq的,2008年7月提供]
基因本体信息
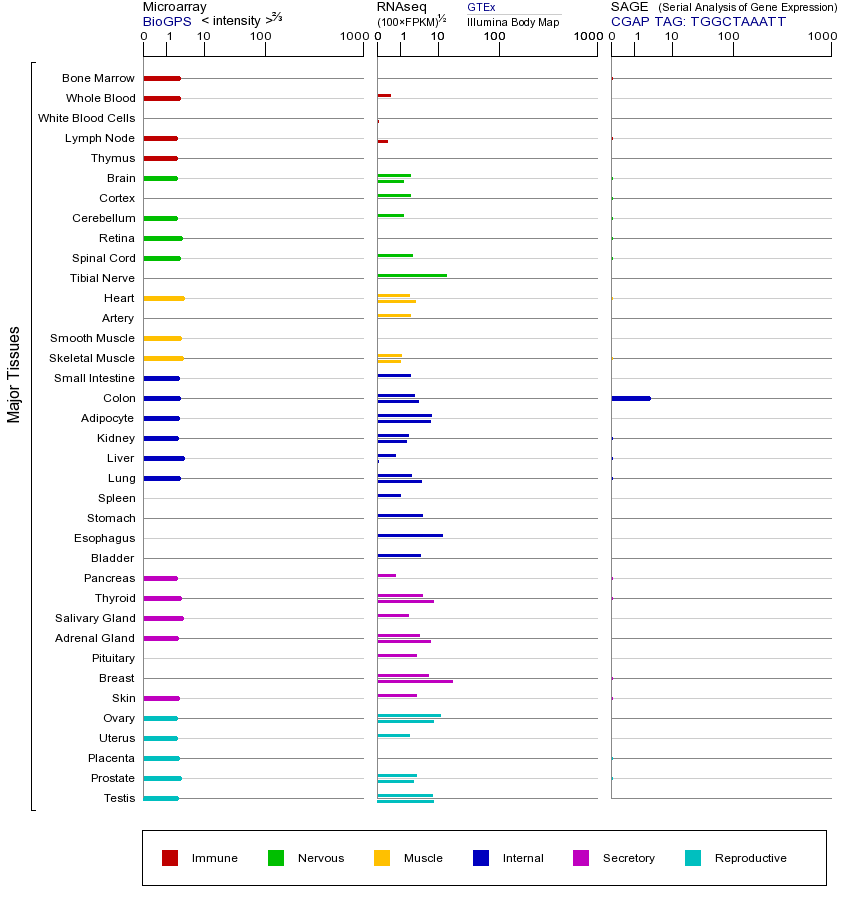
LHCGR基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
LHCGR基因的本体(GO)信息:
| 名称 |
|---|
| 4020 Calcium signaling pathway [PATH:hsa04020] |
| 4080 Neuroactive ligand-receptor interaction [PATH:hsa04080] |
| 4913 Ovarian Steroidogenesis [PATH:hsa04913] |
| 4917 Prolactin signaling pathway [PATH:hsa04917] |
| 名称 |
|---|
| Class A/1 (Rhodopsin-like receptors) |
| G alpha (s) signalling events |
| GPCR downstream signaling |
| GPCR ligand binding |
| Hormone ligand-binding receptors |
| Signal Transduction |
| Signaling by GPCR |
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Familial Testotoxicosis | 0.367524428 | 25 | 2 | BeFree_CTD_human_GAD_ORPHANET_UNIPROT |
| Leydig cell agenesis | 0.360271442 | 10 | 1 | BeFree_CLINVAR_ORPHANET_UNIPROT |
| Polycystic Ovary Syndrome | 0.129544073 | 13 | 4 | BeFree_GAD_GWASCAT |
| Male infertility | 0.122367032 | 2 | 0 | CTD_human_GAD |
| Leydig Cell Hypoplasia | 0.122171535 | 8 | 0 | BeFree_CTD_human |
| Testotoxicosis | 0.121628651 | 6 | 1 | BeFree_ORPHANET |
| Leydig Cell Tumor | 0.120271442 | 3 | 1 | BeFree_CTD_human |
| Disorders of Sex Development | 0.120271442 | 2 | 0 | BeFree_CTD_human |
| 46, XY Disorders of Sex Development | 0.120271442 | 2 | 0 | BeFree_CTD_human |
| Mammary Neoplasms, Experimental | 0.08 | 1 | 0 | RGD |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。