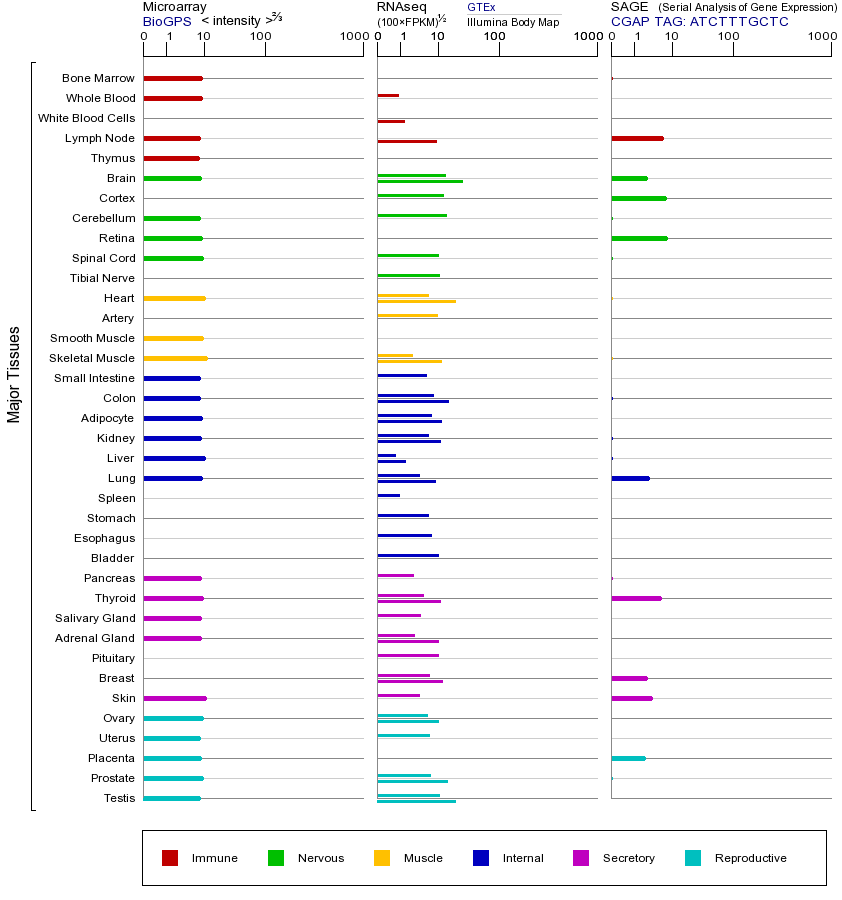
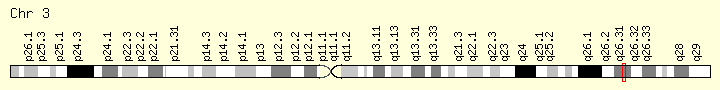NLGN1 (neuroligin 1)
- symbol:
- NLGN1
- locus group:
- protein-coding gene
- location:
- 3q26.31
- gene_family:
- alias symbol:
- KIAA1070
- alias name:
- None
- entrez id:
- 22871
- ensembl gene id:
- ENSG00000169760
- ucsc gene id:
- uc003fio.3
- refseq accession:
- NM_014932
- hgnc_id:
- HGNC:14291
- approved reserved:
- 2001-01-02
NLGN1(neuroligin-1,神经连接蛋白-1)是一种突触细胞黏附分子,属于neuroligin基因家族(包含NLGN1、NLGN2、NLGN3、NLGN4X/Y等成员),该家族共性是通过与突触后膜的神经连接蛋白(neurexin)结合,介导神经元间的黏附与信号传递,参与突触形成、维持及可塑性调控。NLGN1主要表达于兴奋性突触(如谷氨酸能突触),其胞外区含有胆碱酯酶样结构域(负责与neurexin结合),胞内区则通过PDZ结构域与突触支架蛋白(如PSD-95)相互作用,从而锚定突触后致密区(PSD)并招募AMPA/NMDA受体,增强突触信号传导效率。其生物学功能包括促进树突棘形态发生、调节长时程增强(LTP,一种学习记忆相关的突触强化机制)以及维持兴奋性/抑制性突触平衡。 NLGN1突变可能导致功能丧失或异常,与多种神经精神疾病相关。例如,错义突变可能破坏与neurexin的结合能力,导致突触发育缺陷,与自闭症谱系障碍(ASD)和智力障碍相关;而剪切突变可能引发异常亚细胞定位,影响突触可塑性,增加精神分裂症风险。在阿尔茨海默病中,NLGN1表达下降可能加剧突触丢失和认知衰退。 过表达NLGN1会增强兴奋性突触数量和功能,可能改善神经退行性疾病的突触缺陷,但过度表达可能导致兴奋/抑制失衡,诱发癫痫样放电。反之,表达降低会减少树突棘密度、削弱LTP,导致学习记忆障碍。NLGN1还通过调控下游基因(如BDNF、Shank3)影响神经发育,其表达异常可能间接改变突触相关蛋白网络。 该基因家族成员具有高度保守的胞外胆碱酯酶样结构域,但亚细胞定位和功能分化明显:NLGN2/3偏向抑制性突触(GABA能),而NLGN1/4主要参与兴奋性突触。这种分工共同维持神经环路稳态,其异常普遍与神经发育性疾病相关。
This gene encodes a member of a family of neuronal cell surface proteins. Members of this family may act as splice site-specific ligands for beta-neurexins and may be involved in the formation and remodeling of central nervous system synapses. [provided by RefSeq, Jul 2008]
该基因编码一个家族的神经元细胞表面蛋白的成员。这个家庭的成员可能充当的β-neurexins剪接位点特异性配体和可能参与的形成和中枢神经系统突触重塑。 [由RefSeq的,2008年7月提供]
基因本体信息
NLGN1基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
NLGN1基因的本体(GO)信息:
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Autistic Disorder | 0.12827274 | 6 | 0 | BeFree_CTD_human_GAD_LHGDN |
| hearing impairment | 0.12 | 1 | 2 | GWASCAT |
| Hearing Loss, Partial | 0.12 | 1 | 2 | GWASCAT |
| Motion Sickness | 0.12 | 1 | 1 | GWASCAT |
| Status Epilepticus | 0.08 | 1 | 0 | RGD |
| Schizophrenia | 0.002638474 | 2 | 0 | BeFree_GAD |
| Tobacco Use Disorder | 0.002367032 | 1 | 0 | GAD |
| Mental Depression | 0.002367032 | 1 | 0 | GAD |
| Alcoholic Intoxication, Chronic | 0.002367032 | 1 | 0 | GAD |
| Narcolepsy | 0.002367032 | 1 | 0 | GAD |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。