PINK1 (PTEN induced kinase 1)
- symbol:
- PINK1
- locus group:
- protein-coding gene
- location:
- 1p36.12
- gene_family:
- Parkinson disease associated genes
- alias symbol:
- BRPK
- alias name:
- None
- entrez id:
- 65018
- ensembl gene id:
- ENSG00000158828
- ucsc gene id:
- uc001bdm.3
- refseq accession:
- NM_032409
- hgnc_id:
- HGNC:14581
- approved reserved:
- 2001-02-08
PINK1(PTEN诱导激酶1)是一种由PINK1基因编码的线粒体丝氨酸/苏氨酸蛋白激酶,在维持线粒体功能中起核心作用。该基因属于PINK1/Parkin通路,这个家族通过泛素-蛋白酶体系统调控线粒体自噬(mitophagy,即受损线粒体的选择性清除)。PINK1的主要作用位点是线粒体外膜,当线粒体受损导致膜电位下降时,PINK1会在线粒体表面积累并激活Parkin蛋白,进而标记受损线粒体进行降解。PINK1的生物学功能包括监测线粒体健康状态、调节能量代谢、清除功能异常的线粒体以及抑制活性氧(ROS,一种对细胞有害的氧化物质)的产生。PINK1基因突变会导致其激酶活性丧失,最常见的是与早发性帕金森病(一种神经退行性疾病)相关的常染色体隐性突变,这些突变会损害线粒体自噬过程,导致异常线粒体和毒性蛋白的积累,最终引发多巴胺能神经元(负责运动协调的神经细胞)死亡。PINK1过表达可增强线粒体自噬并保护神经元,但过度激活可能导致线粒体过度清除而影响细胞能量供应;而PINK1表达降低会削弱线粒体质量控制,加速神经退行性变。除帕金森病外,PINK1功能异常还与阿尔茨海默病、糖尿病和癌症等疾病相关。PINK1/Parkin基因家族的共性是通过泛素化(给蛋白质添加泛素标记的过程)机制调控线粒体动态平衡,成员间存在功能互补,如Parkin依赖PINK1激活才能发挥作用。目前"PINK1"的中文译名"PTEN诱导激酶1"能准确反映其特性,但部分文献也简称为" Pink1激酶"。
This gene encodes a serine/threonine protein kinase that localizes to mitochondria. It is thought to protect cells from stress-induced mitochondrial dysfunction. Mutations in this gene cause one form of autosomal recessive early-onset Parkinson disease. [provided by RefSeq, Jul 2008]
该基因编码定位于线粒体丝氨酸/苏氨酸蛋白激酶。它被认为是保护从应力诱发的线粒体功能障碍的细胞。突变该基因原因一常染色体隐性遗传早发性帕金森病的形式。 [由RefSeq的,2008年7月提供]
基因本体信息
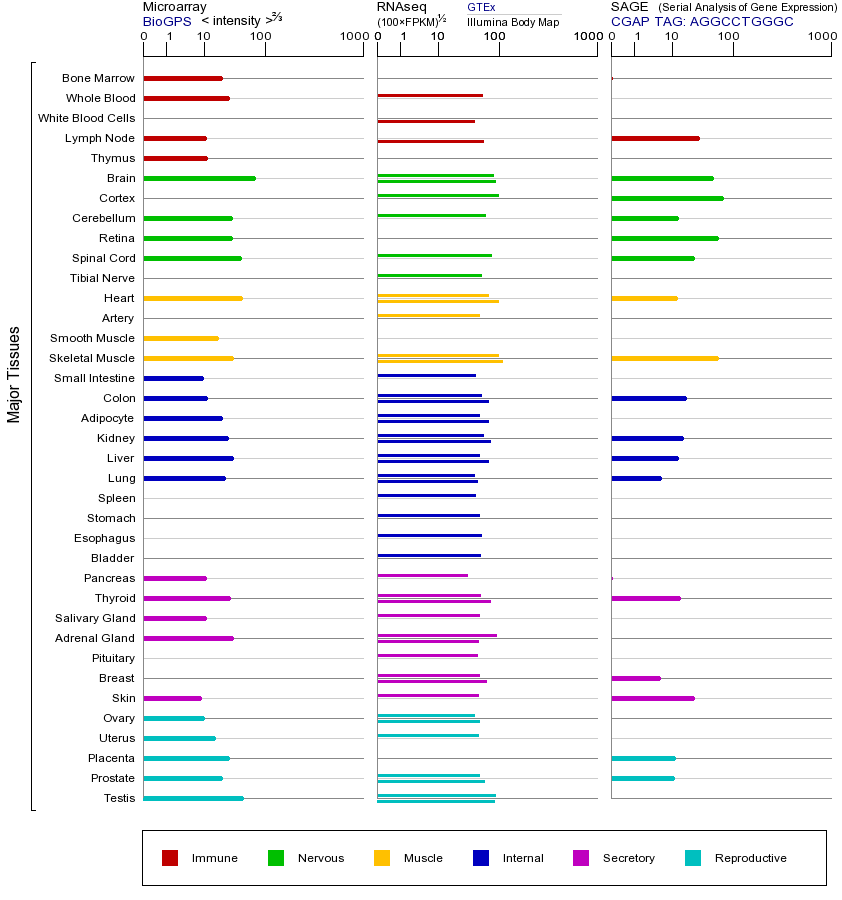
PINK1基因(以及对应的蛋白质)的细胞分布位置:
- 质膜
- 细胞质
- 细胞外
- 高尔基体
- 囊泡
- 细胞骨架
- 内质网
- 细胞核
- 内体
- 溶酶体
- 线粒体
PINK1基因的本体(GO)信息:
| 疾病名称 | 关系值 | NofPmids | NofSnps | 来源 |
| Parkinson Disease 6, Autosomal Recessive Early-Onset | 0.44 | 17 | 3 | CLINVAR_CTD_human_MGD_UNIPROT |
| Parkinson Disease | 0.272130727 | 114 | 3 | BeFree_CTD_human_GAD_LHGDN |
| Parkinsonian Disorders | 0.177698531 | 81 | 2 | BeFree_CTD_human_GAD_LHGDN |
| Neuroblastoma | 0.120814326 | 4 | 0 | BeFree_CTD_human |
| Peripheral Neuropathy | 0.120271442 | 2 | 0 | BeFree_CTD_human |
| Diabetes Mellitus, Non-Insulin-Dependent | 0.005634266 | 2 | 0 | BeFree_GAD_LHGDN |
| Neurodegenerative Disorders | 0.00408156 | 5 | 0 | BeFree_LHGDN |
| Schizophrenia | 0.003810118 | 4 | 0 | BeFree_LHGDN |
| Dementia | 0.003267234 | 3 | 0 | BeFree_LHGDN |
| Autosomal Recessive Parkinsonism | 0.003257302 | 12 | 0 | BeFree |
联系方式
山东省济南市章丘区文博路2号 齐鲁师范学院 genelibs生信实验室
山东省济南市高新区舜华路750号大学科技园北区F座4单元2楼
电话: 0531-88819269
E-mail: product@genelibs.com
微信公众号
关注微信订阅号,实时查看信息,关注医学生物学动态。